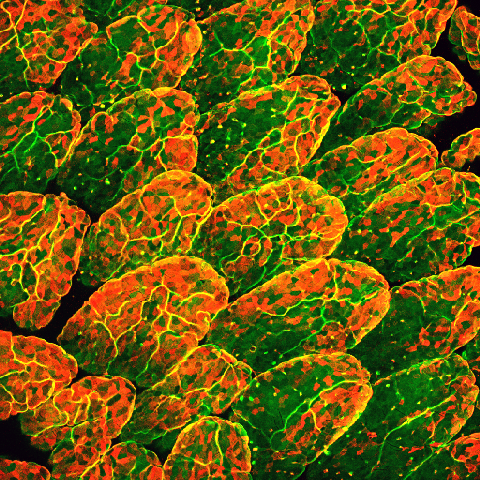
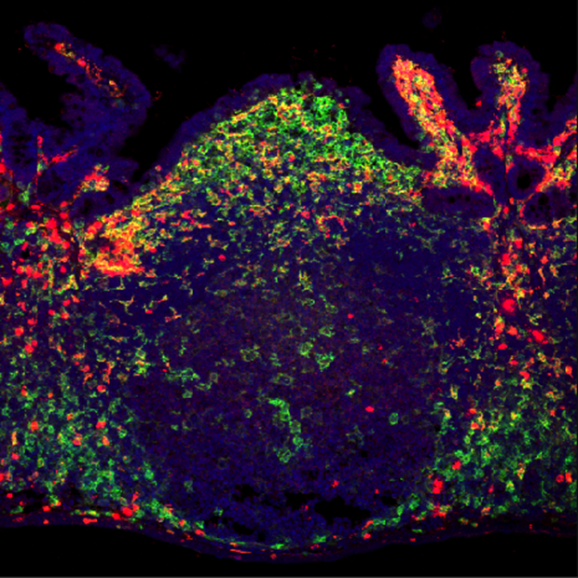
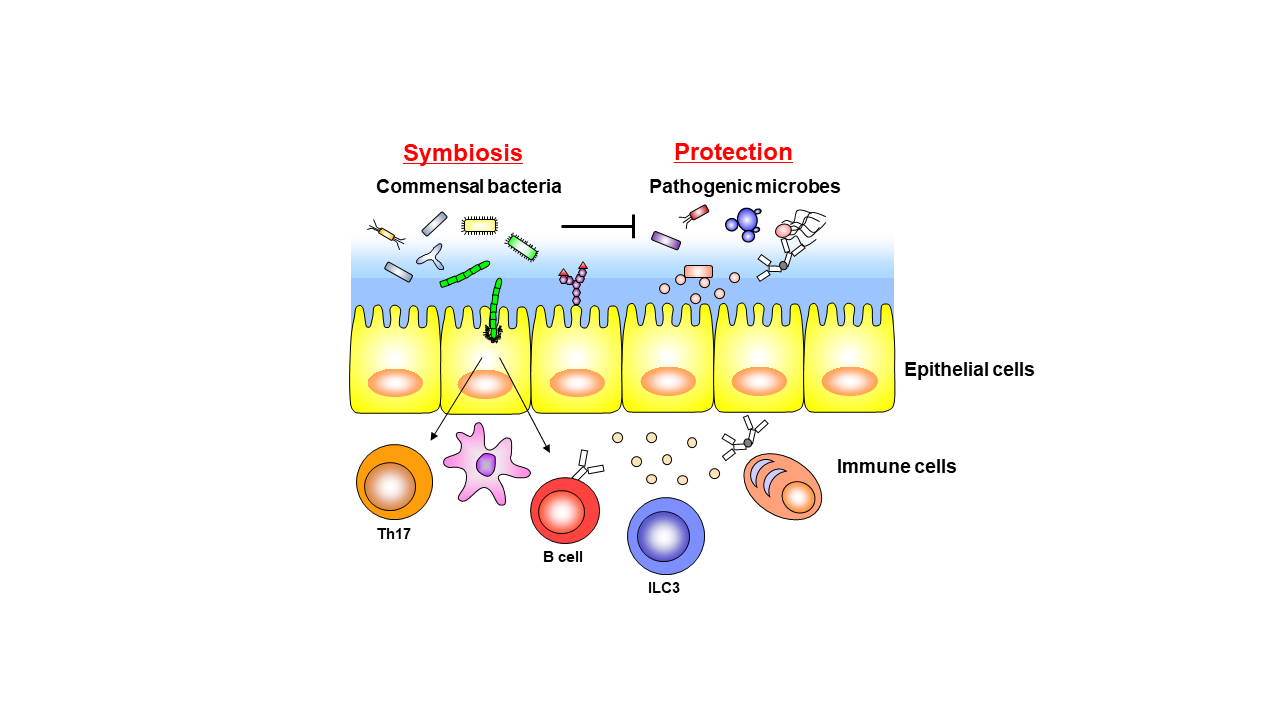
Innumerable microorganisms peacefully colonize human mucosal tissues, including the gut. In particular, many bacteria, fungi, viruses, and parasites residing in the gut exert various effects on the host, such as inducing the differentiation and proliferation of immune cells. It is an attractable biological theme to identify the mechanism of symbiosis between the host and these intestinal microbes. In addition, it has been reported that disruption of the host and microbe symbiotic relationship predisposes the development of various diseases such as infectious diseases, inflammatory bowel disease, allergic diseases, metabolic disorders such as obesity and diabetes, and colon cancer. In contrast to symbiotic responses, host immune cells immediately eliminate pathogenic microbes from the gut. How this "symbiosis" with intestinal microbes and "exclusion" of pathogenic microbes is established is unclear yet. Commensal bacteria residing in the gut play important roles in 1. inhibition of colonization and infection by pathogenic microorganisms in the gut, and 2. induction of the host immune system. Our laboratory discovered that commensal bacteria and the host immune system interact cooperatively to form a first-line defense barrier against pathogen infection. Currently, we conduct daily research with the following research themes through elucidation of the mechanism of "symbiosis" with commensal bacteria and the "elimination" of pathogenic microorganisms.
| Yoshiyuki Goto | Associate Professor |
|---|---|
| Sayaka Hasegawa | Research Promotion Technician |
| Minami Hirayama | Research Promotion Technician |
| Miyuki Yamaguchi | Research Promotion Technician |